Regarding your question:
- You can use both a CSV file and a database. Set the AlphaWell Properties to Labware and create a Data Set Definition for your data if you have a plate barcode or Sample ID.
- When the Biomek transfers liquid from Plate X to Plate Y, all the new data will be transferred to the new plate. You then have two options: you can either generate a CSV file report with the desired format and data, or use Dart Tools to import the data into your database.
- LIMS/LIS will connect to Dart, and the robot will connect to two SQL databases. When you import the plate barcode containing the sample data, it will retrieve the information from the database. Your LIMS software must be integrated with Dart.
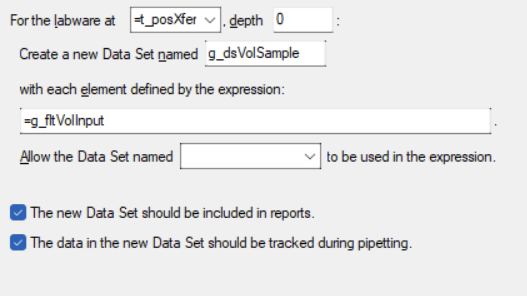
Example of generating a report on the normalization method, including what was transferred and what failed.
Option Explicit
Dim z, i, oFs, oFile, PlateName, FilePath, FileName, FileExtension, sLine, Delimiter
Dim TMP_Well, od_w, rnaconc_w, rnatake_w, addupw_w, addrtreagents_w, wellstatus_w, RNASource_w, DilSource_w,SampleName
Dim Pos(6) ' Declaring the size of the array
' Function to calculate well number
Function WellNumCountingDown(wellNum)
Dim col, row
col = ((wellNum-1) \ 8) + 1
row = (wellNum-1) Mod 8 + 1
WellNumCountingDown = col + ((row-1) * 12)
End Function
' Set positions
Pos(1) = "P7"
Pos(2) = "P11"
Pos(3) = "P12"
Pos(4) = "P15"
Pos(5) = "P20"
Pos(6) = "P23"
' Set File path and delimiter
FilePath = "X:\qPCR\"
Delimiter = ","
' Start the main loop for each plate
For z = 1 To SamplePlate_Num
' Set variables for each plate
FileName =Eval("BRC" & z)' Constructing filename for each plate
FileExtension = ".csv"
' Initialize the FileSystemObject and file for writing
Set oFs = CreateObject("Scripting.FileSystemObject")
Set oFile = oFs.CreateTextFile(FilePath & FileName & FileExtension, True)
' Writing CSV header
sLine = "Well" & Delimiter & "name" & Delimiter & "od" & Delimiter & "rnaconc" & Delimiter & "rnatake" & Delimiter & "addupw" & Delimiter & "addrtreagents" & Delimiter & "wellstatus" & Delimiter & "RNASource" & Delimiter & "DilSource"
oFile.WriteLine sLine
' Process each well
For i = 1 To 96
' The next lines assume the existence of certain methods and properties within your environment.
' Make sure to replace "Positions", "Pipettor.Deck.findlabwareposition", and ".Labware.Datasets" with your actual methods for fetching these data.
addrtreagents_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.addrtreagents(WellNumCountingDown(i))
If addrtreagents_w <> 0 Then
' Access well properties. This part of the code is highly dependent on your environment and objects structure.
SampleName = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.name(WellNumCountingDown(i))
TMP_Well = Positions(Pipettor.Deck.findlabwareposition("P9")).Labware.Datasets.AlphaWellNoZero(WellNumCountingDown(i))
od_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.od(WellNumCountingDown(i))
rnaconc_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.rnaconc(WellNumCountingDown(i))
rnatake_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.rnatake(WellNumCountingDown(i))
addupw_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.addupw(WellNumCountingDown(i))
wellstatus_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.wellstatus(WellNumCountingDown(i))
RNASource_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.RNASource(WellNumCountingDown(i))
DilSource_w = Positions(Pipettor.Deck.findlabwareposition(Pos(z))).Labware.Datasets.DilSource(WellNumCountingDown(i))
' Compile the line for this well's data
sLine = TMP_Well & Delimiter & SampleName & Delimiter & od_w & Delimiter & rnaconc_w & Delimiter & rnatake_w & Delimiter & addupw_w & Delimiter & addrtreagents_w & Delimiter & wellstatus_w & Delimiter & RNASource_w & Delimiter & DilSource_w
' Write the data line to file
oFile.WriteLine sLine
End If
Next ' next well
oFile.Close ' Close the file for the current plate
Next ' next plate
' Cleanup
Set oFs = Nothing
Set oFile = Nothing
I have an attachment method for cDNA and a report generation file. The user will enter the barcode plate (by scanning) into the UI. The method will then import data from a CSV file where the barcode of the plate matches the CSV file name. The CSV file will be located in a specific folder, for example: FileLoc(i) = "X:\cDNA\" & BRC(i) & ".csv". Every time the user enters a plate name that matches a file name in the folder, the data will be automatically imported from that file.(Static Folder called “cDNA”)
Then, when transferring liquid, any value that fails or has a value of 0 will generate a new file in another Static folder called “qPCR”, excluding the samples that failed.
https://www.mediafire.com/file/4c47fstp5nxq9od/Normalization_Biosorts_Services_Plates.bmf/file
https://www.mediafire.com/file/y13yn8t1qqqjgwe/Import_File.csv/file