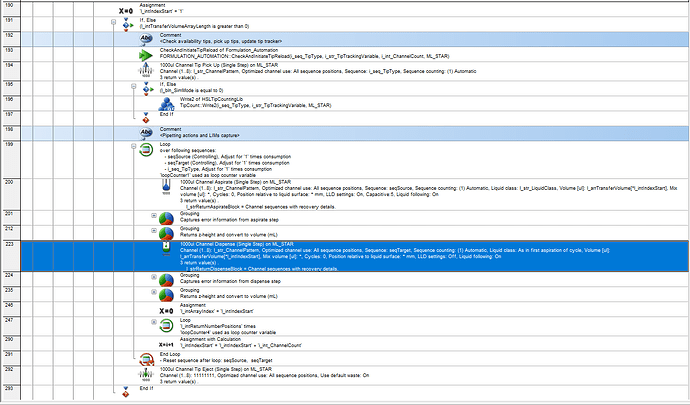
I ran into an interesting problem where the channel useage on a method unexpectedly changed from 8 to 6 on the second loop of an aspirate and dispense loop and I’m curious if anyone else has experienced this or has some insight to why it might have occurred.
I have a method, that has run without error/issue for a few weeks now. The method looks at the source sequence and determines the number of tips needed to complete the sequence with reusing tips. It then picks up the tips (almost always 8 total) and draws from the 8 positions of an RGT trough before dispensing into the target labware. However, on a recent run I observed the STARlet pick up 8 tips but aspirate/dispense with 6 channels. On the next set of eight, the RGT positions became misaligned with the rest of the sequences and eventually ended in a fatal error with the method being aborted.
I’m at a loss at why this occurred since the code did not change during this time and had worked perfectly prior. Upon receiving this error, I looked at the code but could not determine a cause, and decided to run the method again without making any changes. This rerun proceeded as expected and the issue was not seen… ![]() To add to this, the code that controls this aspirate/dispense loop was run twice during the errored run. The first loop which pipets buffer 1, complete as intended (8 channels, 8 aspirates/dispense) but on the second pass of the loop it use 6 channels during the aspirates/dispenses for buffer 2 and encounter this error.
To add to this, the code that controls this aspirate/dispense loop was run twice during the errored run. The first loop which pipets buffer 1, complete as intended (8 channels, 8 aspirates/dispense) but on the second pass of the loop it use 6 channels during the aspirates/dispenses for buffer 2 and encounter this error.
I’m uncertain why this issue occurred on this single run and not any others, and furthermore that it occurred on the second pass of the loop and not the first…
I’ve attached snippets of the loop that controls the aspirate/dispense, the trace file, and .csv file that the steps are read in from. Hopefully someone can provide guidance for this issue or see something I overlooked.
This loop is contained within another loop to process multiple buffers with the same code. The instrument picked up 8 tips as instructed, but then aspirated on 6 channels in remaining aspirate and dispenses. Obviously not all the code is shown due to limits in image size. More info here can be provided if needed.
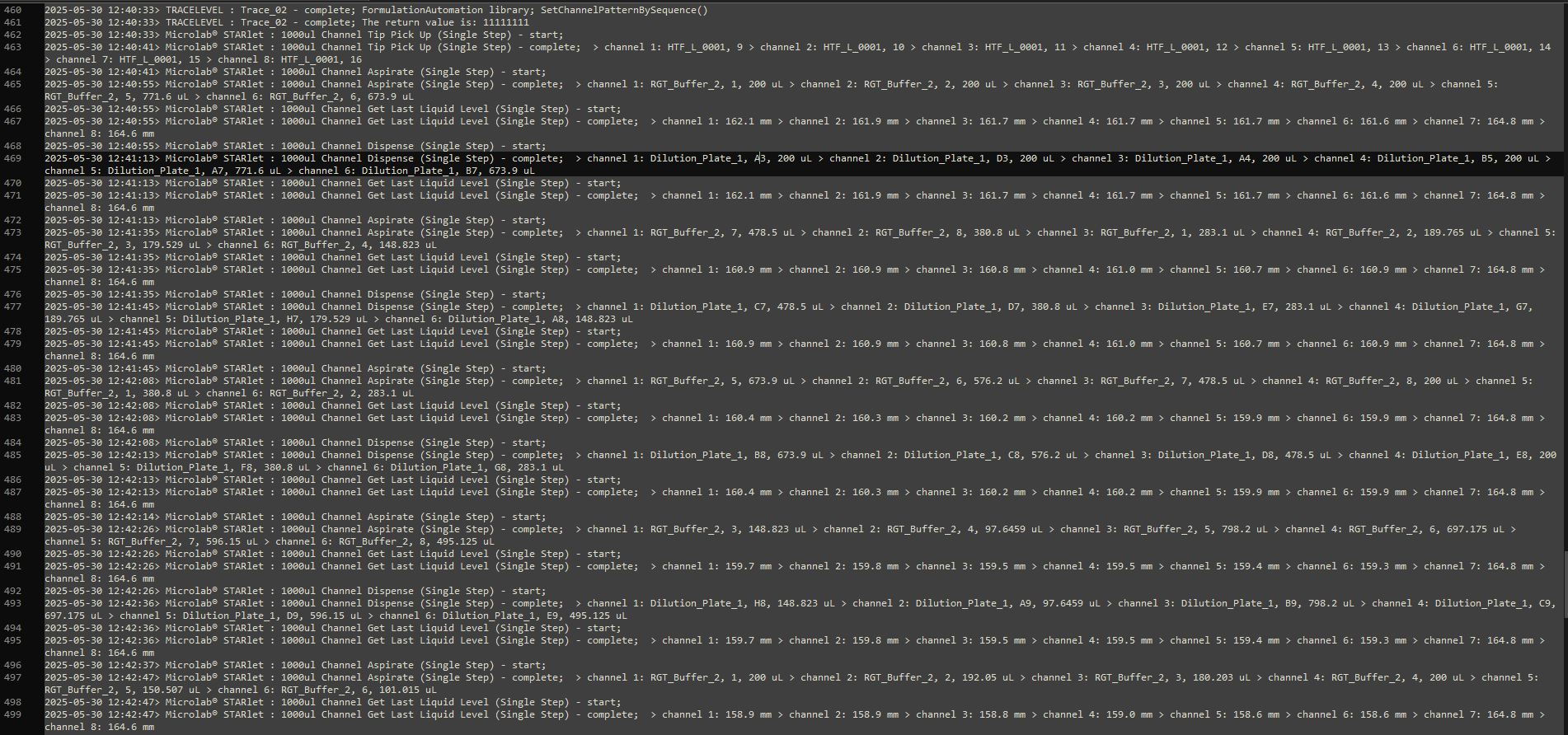
This is the trace file for the above loop when the error occurred. On line 473 we can see that channel 1 used the unused positions (RGT_Buffer_2, 7 & RGT_Buffer_2, 8) from the last aspirate instead of position 1 & 2 but the the correct volume of 478.5uL. This is where the position sequences became misaligned. This continued until an index for an array position was request that did not exist resulting in the fatal error.
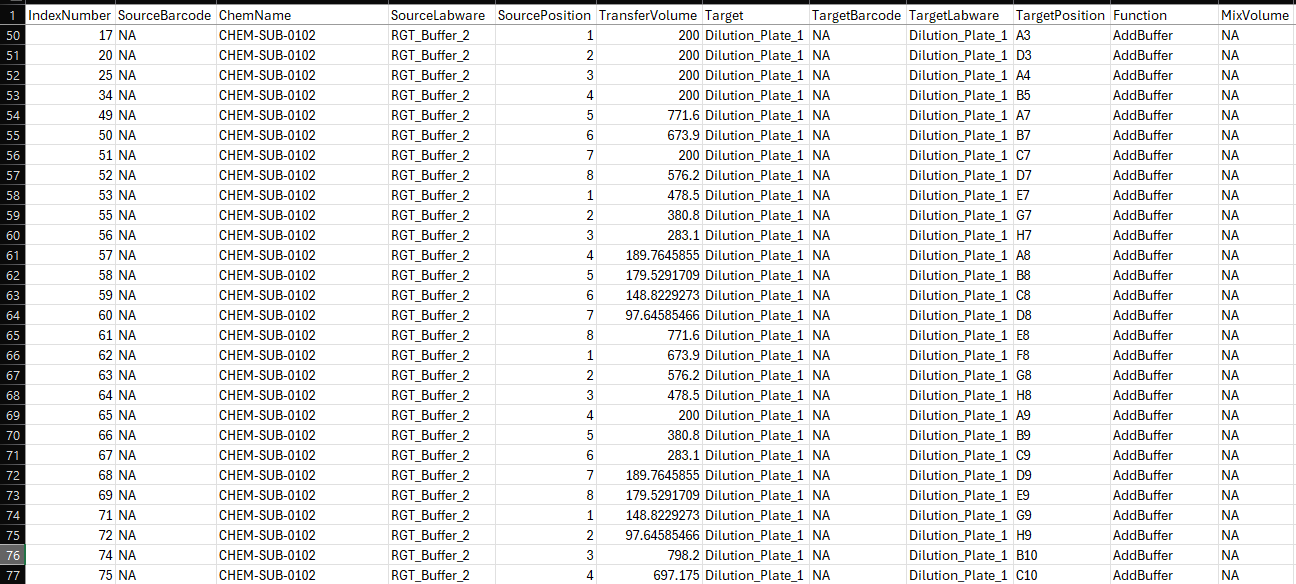
This is the CSV file read in to instruct the STARlet of the pipet steps. We can see that a RGT_Buffer_2 positions of 1-8 is request and should be processed in a group since we have 8 channels, but that is not what occurred. If we look at the volumes and example above in the trace file. The misalignment that occurred in the trace file (line 473) is a combination of index number 51 of csv that uses source position 7 and index number 53’s transfer volume.
I’m a bit concerned at the inability to reproduce or find the root cause of this issue since it appeared randomly in a working method and disappeared on a rerun. If this had occurred during the use of precious samples, those samples would have unrecoverable as well.
Hopefully, someone can see something that I might have overlooked and point me in a direction to prevent this from happening again. If additional information is needed, please let me know and I’ll provide what I can.
-EH421