Hi all,
I’ve recently automated DNA cleanup on Hamilton Start using magnetic beads - many thanks to this forum for help support! The problem now is that manual purifiation repeatedly yields 10-30% more DNA from the same input compared to the robot. I can’t see beads in the waste so unless they are somehow stuck inside the black tips (which I can’t see through) I have no explanation as to why the yields are lower? Has anyone experienced (and hopefully resolved) such an issue? or has recommendations for troublshooting/optimization?
Have you tried repeating the elution step after the robot ?
Typically, I’ve seen situations where the recovery off the beads requires a second elution,
if you do this manually, you could find the missing 10-30% yield
Thanks @Optimize - will give it a go later today ![]()
Could be quite a few reasons why you have lower yield on the robot.
Are you using the same exact protocol for each comparison?
How do the beads get added on the robot? Are they staying well suspended or are they sometimes sitting for a period of time and perhaps separating a bit? PEG concentration will directly affect the size of DNA that will bind to the beads. Have you run a gel to see if you’re losing any DNA of a certain size?
Are you using the same magnet?
Are incubation times the same (binding, elution, and beads pelleting incubations)?
Hi @jnecr . My model system is 30uL of 3kbp plasmid at 1ng/uL + 30uL of beads. Because of the 3kb size, I don’t think size-related losses are of concern. Plasmid is aliquoted equally for robot and human, after that the two workflows run independently but use all the same reagents, so beads are added by the robot to robot’s samples and by human to human’s samples. The magnets are different but I’m not sure if this is worth addressing? if it’s better with “human” magnet I can’t really change this for the robot. If only to establish the root cause , then yes.
Same plate labware for each protocol? There are a lot of magnet options out there and they will perform differently. Either with regards to holding the pellet while the robot removes supernatant or creating the pellet to begin with. You could do some tests with no DNA and collect the supernatant from the robot in a tube of some sort and pellet anything in that tube on a magnet to see if you’re losing beads there.
60uL volume should be pretty easy to pellet for most magnets, so perhaps some beads are getting dislodged during supernatant removal steps, that’s probably where I would start.
Are you looking at the beads after the drying step prior to elution? They may be overdried which IIRC can reduce recovery.
99% of yield problems with nucleic acid purification come down to the binding or elution steps.
Are you adding the correct amount of beads? Particularly important for cleanups but bead “over saturation” is rarely a thing for normal extractions.
Are you mixing well enough? My rule of thumb is Vortexing > tipmixing > orbital shaker. You can’t vortex on a robot, but you can tipmix for extended durations.
Are the beads dry prior to elution? EtOH carryover can greatly affect quantification on OD-based methods, and will negatively impact downstream work, especially PCR. A second aspiration 30s-60s delayed from the initial liquid waste removal can greatly assist in drying; besides that, 3-10m is average drying time at ambient temp. Relevent to @mnewsom 's comment, my rule of thumb is “Shiny, not cracked” for how the pellet/ring should look.
Is your elution efficient? Heated elution can assist in yield recovery if applicable. Similar to binding, mixing efficiency and precision of volume transfer will be essential.
Finally, it’s worth stating that there is loss when re-suspending beads post-dry, typically of around 3-5µL of volume will be sequestered by the bead pellet and become unrecoverable. Understand this lost volume and add ~2-5µL extra volume to your elution dispense to recover a more consistent volume of purified, extracted product.
Great input also want to add that sometimes that first binding step is the most important one and may require more time. In addition, if you do use an orbital shake, 3.0 mm is the way to go and IMO as good as tip mixing despite it potentially taking longer. in addition, if you’re heating make sure you get proper well coverage. Bottom contact alone won’t be enough. And yes the magnet can absolutely matter. Settling time is a big concern. Finally some manufacturers have specific bead to final elution buffer ratios. Find those out if you can.
edit: just remembered/saw your fragment size was pretty large. Leaving up for others though.
Yeah beads are pretty difficult to pipette and you’re likely underdispensing to some degree which would cut the smaller fragments of your product.
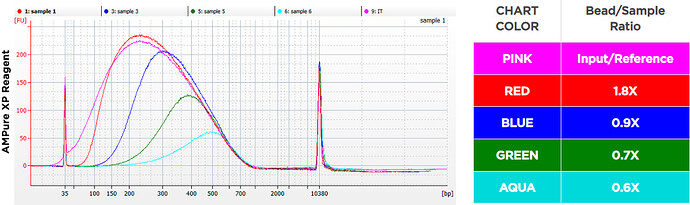
See https://www.beckman.com/reagents/genomic/cleanup-and-size-selection/pcr/bead-ratio
Specifically:
Thanks everyone for great input! I tried to address most comment below but wihtout tagging individuals due to some overlap in themes between comments, hope this is not an issue.
I’ve done some further testing and it appears that at least the elution step is suboptimal - manual resuspending of the robot-produced washed beads improves yield. This is strange given that elution on the robot is carried out by shaking in Hamilton heat shaker for 60 sec at 1700 rpm (at room temp). I’m not sure the amplitude (there was a note about 3 mm) and don’t think this is something that can be programmed/controled from Venus.
Other steps migth also be problematic but will take more time to access. Beads are resuspended by pipetting up and down at low speed. heat shaker does not seem to help much and occasionally the volumes are high enough to risk cross-well carryover if shaking
I use Qubit dsDNA HS for DNA quantification so ethanol carryover (if any) should not impact the reading , although it may impact elution efficiency. Already after 2 min the pellets look dry by eye, not shiny. We previosuly compared over- vs under- vs “normal”- drying of beads in the manual cleanup and found that overdrying is not a problem while underdrying may be, depending how “wet” they are. Still, some fellow scientists were not convinced with this and continue to avoid overdrying. For the manual process with settled on something like 3 minutes.
Finally, I tried manual cleanup using the same palsticware and magnets as used on the robot. In this case yields were much closer between human (me) and robot but mostly becasue human yields got lower - sort of good news, bad news ![]() Ultimately , if the yields in manual procedure are better than on the robot, regardless of the reason, switching to the robot will be a hard sell to the team, because high yields are important for us.
Ultimately , if the yields in manual procedure are better than on the robot, regardless of the reason, switching to the robot will be a hard sell to the team, because high yields are important for us.
Plan for now is to repeat the process with few manual intervensions at varios points, staring from supplying the same beads+DNA mixture to both human and robot (currently robot delivers beads) and then follow up step by step in attempt to identify where efficiency diverges / takes main hit. As I mentioned at the start , beads are not being lost in waste , although they might be trapped inside the black non-transparent tip (I can’t see)
Try to elute for 5 min, 1 min seems to be rather short to me!
Hamilton offers clear tips in addition to the black, conductive tips. It can be very helpful for troubleshooting purposes to check for residual liquid, etc. Follow up with your local sales and support team to see if they could provide a sleeve of clear tips to assist in your investigation and let me know if you need help getting in touch with anyone.
I’ve done some… uhh… unplanned “tests” that involved leaving dry beads on a liquid handler overnight and it’s never been a problem. There’s a lot of superstition when it comes to mag beads, they are actually more resilient than most think.
What kind of magnet do you have for your Hamilton? I like Alpaqua magnets, Olaf has a magnet for every purpose and he is infinitely knowledgeable about magnets and bead separations.
In my experience the first magnetic binding step on the magnet is crucial. I doubled the time for a process in the past (along with more though mixing) and yields shot through the roof. Just something else to consider.
Actually, we’ve been down a similar road, albeit with a different application (veterinary samples).
The issue is very likely the suspension (and dissipation) of the beads for the complete time. Even with constant mixing settings, it is not guaranteed, that the beads are actually well mixed. I found this whitepaper quite helpful, even though it may be a little nerdy: Optimization of orbital mixing parameters in lab automation (qinstruments.com)
What resolved the issue for us in the end, was switching to an integrated magnetic particle processor (there are several such devices on the market). With the direct mixing, the result in our case regularly surpasses the manual method (spin columns). I don’t know how applicable it is to your application. Our solution was recently published here: Animal and Veterinary Sciences | Applications | Hamilton Company
Possibly just one more thing to test, but one solution to this is to split the bead volume. For example, you could add half your volume of beads and mix at high rpm before reducing speeds and reducing mix volume. This has worked for me in the past with things like 10mL cfDNA extractions.
Bind longer and warm the elution.
Question: do you really need that extra 10-30% yield, does it impact on your downstream protocol (i.e. not getting enough, or not getting enough diverse fragments)?
I only query because as automation scientists, we can quite often go down the pursuit of perfection, trying to make everything match the manual or be even better. When you take a step back, sometimes you realise that the benefits that automating it bring can justify a 10% reduction.
Apologies if this isn’t a helpful comment. However it’s a trap I’ve fallen into many times, when I’ve then realised it actually doesn’t matter what the yield is, because overall it does what it needs to do.
Hi @ClaireB
sadly, on this occasion material recovery is very improtant hence my obsession with optimization. Although it’s true that I’d like to make it perfect regardless ![]()