Hey everyone,
FYI the new software update for the Prep has dropped, and adds some cool features/functionality. Details and instructions at this link:
Hi Michael, do you have a list of version updates/history from 1.0 to 3.1?
Thanks!
Hi @Sean_M,
The Prep has come a long way since version 1.0 with 3.2.0 now being the latest release (released at the end of July). The 3.2.0 release adds support for the new Prep CAP HEPA/UV system, support for field verifications, adds drag and drop re-ordering of procotol steps, overhauls the contextual help system, and adds the first of some future visual polish changes.
There have been hundreds of other features and improvements that have been made since 1.0.12 (the initial release). While the change file is not currently posted on the Hamilton Prep landing page, the Lab Solutions Product Support group can quickly answer any specific changes you might have about changes from the past.
Hello everyone!
Sorry, I couldn’t start a new topic for my question.
I am looking for ways to improve some traceability steps in our Microprep equipment. Our idea is as follows:
Protocol: Two plate-to-plate transfers (extracted samples → new plate with the supernatant for subsequent drying).
Before the transfer, I would like to have a way to verify the process via a file or confirmation using the barcodes on the side of the plates. We have an external barcode reader attached to perform the procedure and place the plates on the equipment desk.
We noticed that there is a step for “barcode reading”, but the information is only available at the end, when the report is generated after the pipetting steps.
The process would look something like this:
- Read the file containing the expected barcodes (a list generated by another device).
- Open a window for manual plate scanning and desk positioning.
- Verify the scanned barcode against the expected one.
- Proceed to the pipetting steps.
Does anyone have any suggestions or experience regarding this issue?
Hi @rodrigoschuch,
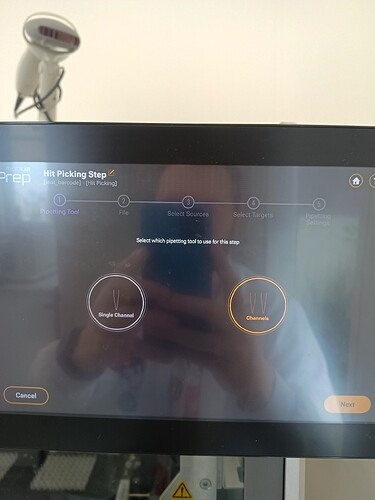
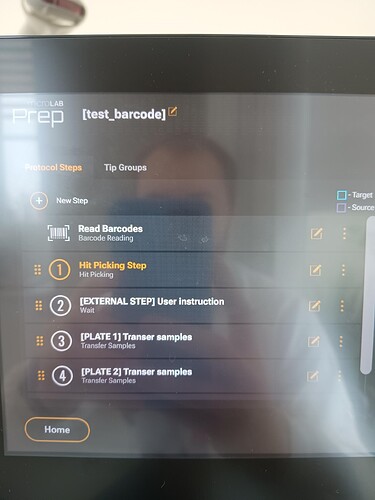
I’m not sure if this is completely what you’re looking for, but the “Read Barcodes” step can be used in conjunction with a “Hit Picking” (HP) step or “Reagent From File” (RFF) step to further drive where your protocol will pipette.
At run time (as I’m sure you’ve seen), the user will be prompted to scan barcodes. These barcodes are then checked (if specified in the HP or RFF steps) against the supplied worklist file for the same barcodes and will use where they were scanned to perform the pipetting. If the barcodes scanned are not found in the provided file, then no pipetting is performed and the protocol will abort.
Have you tried using these “Read Barcodes” in conjunction with either of these steps as a possible solution?
Hello @mrthorne
Thanks for your help. I followed your advice and tested using the combined functions of Hit Picking with barcode reading. What I observed is that the Hit Picking function is not enabled for transfers with the 8-channel multiprobe we have, which would make the protocol very slow.
Ahh, I wasn’t aware you were doing this with the 8MPH. Right, as you saw, hitpicking is only available with the independent channels at the moment (as of v3.2.2). The worklist input file becomes a bit unruly to validate for use with the MPH so that functionality is still slated for a future release.
The wait step you have in your screenshot isn’t a bad path to take for a prompt to your operator, but it isn’t as elegant and programatic as being able to read the barcodes and then validate them against the worklist input file.
Coming in the 3.3.0 release will be ability to export your results in either CSV or JSON (instead of only the PDF) and for the entire run or on a per step basis. While these reports will still only be available at the end, it would at least allow you to programmatically verify what was used.
Thank you for your support.
We might consider an intermediate offline solution for this tracking, something like a Python script that generates a file with some instructions in figures (following the same logic we thought of with Venus).