Hello everyone,
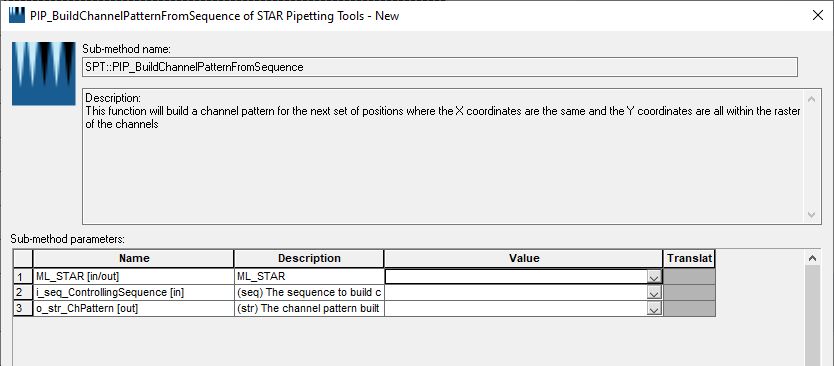
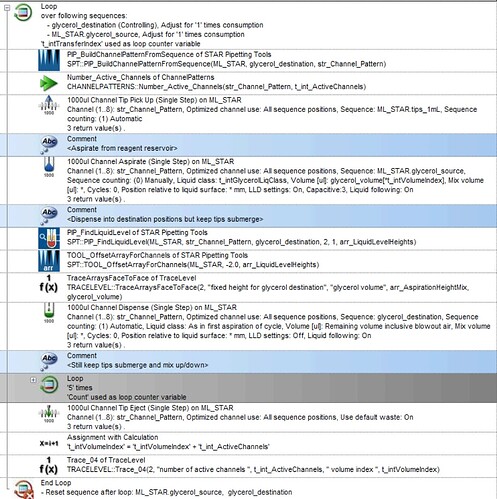
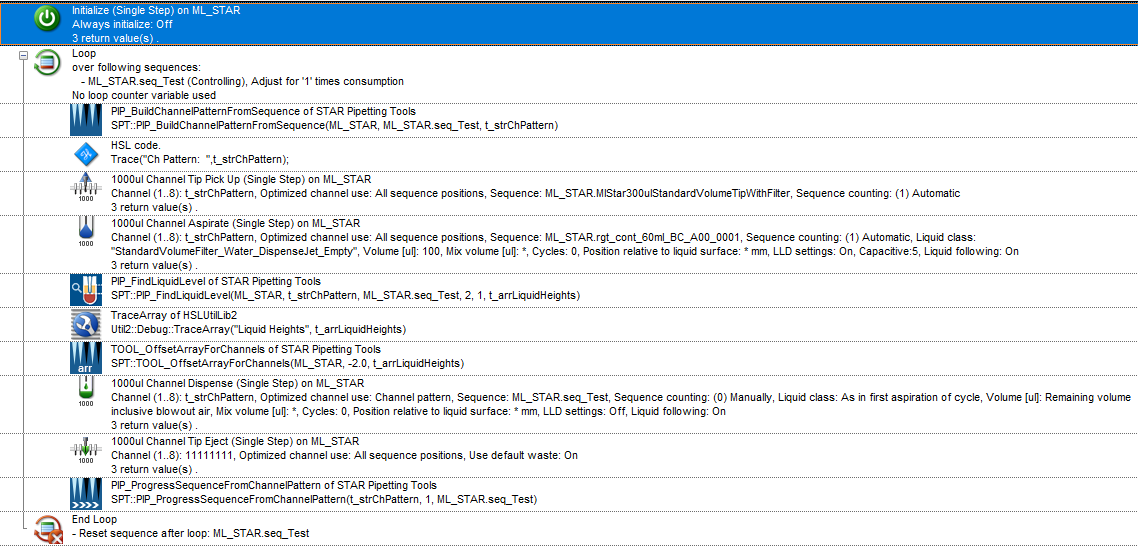
I am working on a protocol that involves a step where I transfer a reagent from a reservoir to some conical tubes. I am utilizing the HighVolumeHelper library to split any large volume of reagent transfer and the STAR Pipetting Tools library to subsequently mix the samples at different fixed heights. However, there seems to be a problem with how the STAR Tool processes the output sequence from the HighVolumeHelper Library.
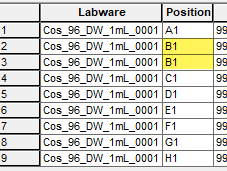
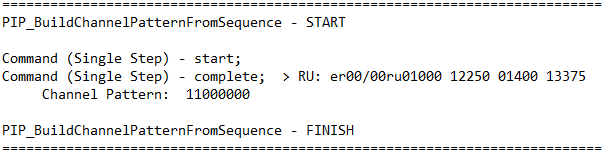
The HighVolumeHelper Library can successfully split the large sequence and update the volume array and destination source accordingly (see below). For example, alpaqua_50_mL_0001, PositionID : A1 is duplicated twice to account for splitting a large volume of 1212 uL.
2024-06-28 13:51:59> TRACELEVEL : TraceArray - start; --------------------------
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [1]: 134.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [2]: 606.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [3]: 606.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [4]: 105.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [5]: 292.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [6]: 145.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [7]: 645.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [8]: 120.0
2024-06-28 13:51:59> TRACELEVEL : TraceArray - progress; Glycerol_volume [9]: 210.0
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Name: l_seqTemp
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Current: 1
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Count: 9
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Total: 9
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Max: 9
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; Used: 0
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: output_1, PositionID : 1
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: alpaqua_50_mL_0001, PositionID : A1
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: alpaqua_50_mL_0001, PositionID : A1
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: output_1, PositionID : 3
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: alpaqua_50_mL_0001, PositionID : A2
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: output_1, PositionID : 5
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: alpaqua_50_mL_0001, PositionID : A3
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: output_1, PositionID : 7
2024-06-28 13:51:59> TRACELEVEL : TraceSequence - progress; LabwareID: output_1, PositionID : 8
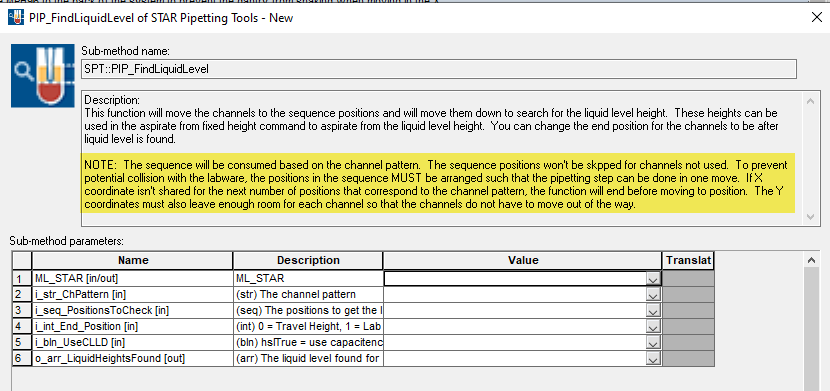
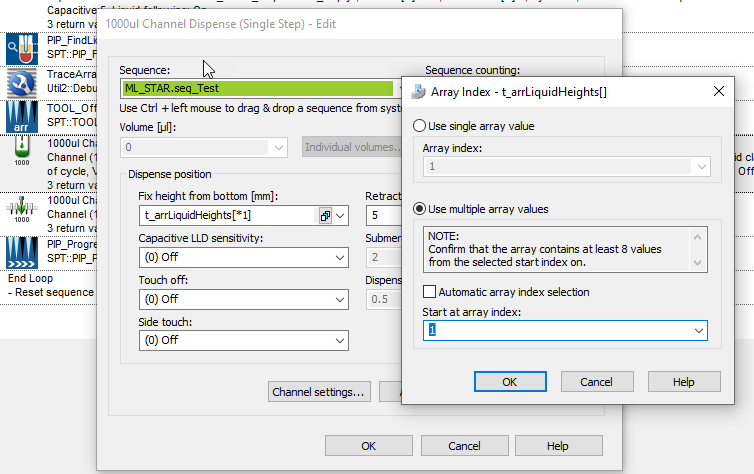
However, when I am using the SPT::PIP_FindLiquidLevel function (for example) to build the pipetting height array based on this new sequence, it seems to only build the array based on unique positions, ignoring the duplicated ones.
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [1]: 18.0 glycerol volume [1]: 134.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [2]: 18.0 glycerol volume [2]: 145.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [3]: 18.0 glycerol volume [3]: 105.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [4]: 18.0 glycerol volume [4]: 120.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [5]: 18.0 glycerol volume [5]: 210.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [6]: 18.0 glycerol volume [6]: 606.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [7]: 18.0 glycerol volume [7]: 606.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; liquid level for glycerol destination [8]: 18.0 glycerol volume [8]: 292.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - progress; glycerol volume [9]: 645.0
2024-06-28 13:52:02> TRACELEVEL : TraceArraysFaceToFace - complete; ----------------------------------------------------------------------------
Because of this, I am receiving an error where the array index is out of bound since the two arrays are not equal, resulting in the step not working. ![]()
I was wondering if there is a way around this limitation. I could split the reagent transfer step and mixing step, but that would increase time and use up more tips.
Any help greatly appreciated,
Nat