Hi All. I wanted to contribute some examples for how to label liquids with the API version 2.14, as the API documentation for Labeling Liquids is a little sparse.
Opentrons gives the examples of:
greenWater = protocol.define_liquid(
name="Green water",
description="Green colored water for demo",
display_color="#00FF00",
)
blueWater = protocol.define_liquid(
name="Blue water",
description="Blue colored water for demo",
display_color="#0000FF",
)
followed by:
well_plate["A1"].load_liquid(liquid=greenWater, volume=50)
well_plate["A2"].load_liquid(greenWater, volume=50)
well_plate["B1"].load_liquid(blueWater, volume=50)
well_plate["B2"].load_liquid(blueWater, volume=50)
reservoir["A1"].load_liquid(greenWater, volume=200)
reservoir["A2"].load_liquid(blueWater, volume=200)
This is great and works fine for loading individual wells/reservoirs. But what I wanted to do was label multiple wells, ideally matching the number of plate columns that our end user would process.
My initial thought process was to try and use by using the Labware.rows_by_name() and Labware.columns_by_name() , with an if else statement that would control how many columns I labeled, based on the number of samples I processed. However, I found and confirmed with Opentrons support that the load_liquid command will only accept individual wells.
With some help from Opentrons, I instead was able use a for loop to accomplish labeling. You can see the code below (with some extra information to give context):
import math
import json
from opentrons import protocol_api, types #needed for trying to set specific pipette movements before pipetting
from opentrons import simulate
def get_values(*names):
_all_values = json.loads("""{"temp_deck":"temperature module gen2",
"pipette_type":"p20_multi_gen2",
"pipette_mount":"right",
"pipette_type_2":"p300_multi_gen2",
"pipette_mount_2":"left",
"sample_number":17,
"site":"Redacted",
"sample_volume":4,
"master_mix_volume":6.2,
"set_temperature":4}""")
return [_all_values[n] for n in names]
metadata = {
'protocolName': 'Kapa_Illumina Library qPCR V2 Step 2',
'author': 'Redacted',
'source': 'Kapa KR0405 v9.17 Protocol',
'apiLevel': '2.14', 'softwareLevel': '6.3.1'
}
def run(protocol_context):
[temp_deck, pipette_type, pipette_mount, pipette_type_2, pipette_mount_2, sample_number, site, sample_volume,
master_mix_volume, set_temperature] = get_values( # noqa: F821
"temp_deck", "pipette_type", "pipette_mount", "pipette_type_2", "pipette_mount_2", "sample_number", "site",
"sample_volume", "master_mix_volume", "set_temperature"
)
# Load Temperature deck in Slot 10
temp_deck = protocol_context.load_module(temp_deck, '10')
# Populate temp_deck with 96 well PCR Reagent Plate
temp_plate = temp_deck.load_labware(
'biorad_96_wellplate_200ul_pcr')
# Load 384 well plate in Slot 1
qPCR_plate = protocol_context.load_labware(
'biorad_384_wellplate_50ul', '1', 'qPCR plate')
# Load 1-10K_1-20K 96 well dilution plate in Slot 3
dilution_10k_20k_plate = protocol_context.load_labware(
'biorad_96_wellplate_200ul_pcr', '3', 'dilution 10k plate')
# Use only 20 uL tips per sample in this protocol, tip rack goes in slots 4-9 and 11
total_tips = (96*7) # Max Number of Tips available given open deck spaces.
tiprack_num = math.ceil(total_tips/96)
slots = ['4', '5', '6', '7', '8', '9', '11'][:tiprack_num]
tip_name = 'opentrons_96_tiprack_20ul'
tipracks = [protocol_context.load_labware(tip_name, slot) for slot in slots]
# Telling Pippete Mount (right_pipette, in this case 20 ul multichannel) to use 20 uL tips
pipette = protocol_context.load_instrument(
pipette_type, pipette_mount, tip_racks=tipracks)
# Set Speed of Z Axis
pipette.default_speed = 100 #Used to use protocol_context.max_speeds['Z'] = 20 to Set Speed of Z Axis
#Math to make loops work for samples variables
col_num = math.ceil(sample_number/8)# IE the total # columns you will be processing.
output_standards = [col for col in qPCR_plate.rows()[1][18:21:1]]#Start at Row B, 19th column (18 is used since Python is 0 based), and proceed until you reach column 21. It will stop when reaching column 22 (21 set here since Python is 0 based) . In this case, MM and standards go into B19, B20, and B21.
# Define Reagent Source Columns
master_mix_col_1 = temp_plate.wells()[0]#IE in Column 1, this is the MasterMix to use for the 1:10K Dilutions
master_mix_col_2 = temp_plate.wells()[8] # IE in Column 2, this is the MasterMix to use for the 1:20K Dilutions and for the Standards/NTCs
standards_col_4 = temp_plate.wells()[32]# IE in **Column 5** Includes Standards in A5-F5; NTCs are in G5 and H5.
#Math for User Deck Preparation
load_tips = (sample_number*12)+48
load_tip_boxes = math.ceil(load_tips/96)
col_1_MM = math.ceil((((col_num*8)*3)*master_mix_volume)/8+20)
col_2_MM = math.ceil(((((col_num*8)*3)+24)*master_mix_volume)/8+20)
col_4_STDs = (sample_volume*3)+20
# Define Liquids to Display during loading prompts
blue10KSample = protocol_context.define_liquid(
name="10K Sample Volume",
description="Blue Colored Initial Sample Volume",
display_color="#7AC5CD",
)
yellow20KSample = protocol_context.define_liquid(
name="Sample Volume",
description="Yellow Colored Initial Sample Volume",
display_color="#FCFF4D",
)
greenMM = protocol_context.define_liquid(
name="MasterMix",
description="Green Colored Initial MasterMix Volume",
display_color="#66cc66",
)
crimsonStandards = protocol_context.define_liquid(
name="Standards",
description="Crimson Colored Initial MasterMix Volume",
display_color="#dc143c",
)
NTCWater = protocol_context.define_liquid(
name="NTC Water",
description="Bronze Colored NTC Water Volume",
display_color="#cd7f32",
)
NTC10mMTris = protocol_context.define_liquid(
name="NTC Tris",
description="Pink Colored NTC 10 mM Tris HCl Volume",
display_color="#E47FBA",
)
#Define Initial Dilution Plate Volumes in uL for Visual Display
Dilution_Well_Vol_10K = 50
Dilution_Well_Vol_20K = 100
#Labeling MasterMix Reagent Plate Wells with Liquids for User Prompts
#label MasterMix Columns 1-2, this is always the same no matter the number of samples, so I hardcode it here.
for i in range (0,8):
temp_plate.wells()[i].load_liquid(liquid=greenMM, volume=col_1_MM)
for i in range (8,16):
temp_plate.wells()[i].load_liquid(liquid=greenMM, volume=col_2_MM)
#Label standard. this is always the same no matter the number of samples, so I hardcode it here.
for i in range (32,38):
temp_plate.wells()[i].load_liquid(liquid=crimsonStandards, volume=col_4_STDs)
#Label NTCs. this is always the same no matter the number of samples, so I hardcode it here.
temp_plate["G5"].load_liquid(liquid=NTCWater, volume=col_4_STDs)
temp_plate["H5"].load_liquid(liquid=NTC10mMTris, volume=col_4_STDs)
#Label 10K Samples
if sample_number <= 48: #the Max Number of samples we process is 48. Any more than that will present the end user with an error message to reload the script with less samples.
for i in range (0,sample_number):
dilution_10k_20k_plate.wells()[i].load_liquid(liquid=blue10KSample, volume=Dilution_Well_Vol_10K)
else:
protocol_context.comment("You have selected >48 samples. Please Enter in only <=48 samples. Reload the protocol script and correct the sample number!")
#pass would also work if we don't want a user prompt there and just want to continue the script.
#pass
#Label 20K Samples
if sample_number <= 48:
for i in range (48,(sample_number+48)):
dilution_10k_20k_plate.wells()[i].load_liquid(liquid=yellow20KSample, volume=Dilution_Well_Vol_20K)
else:
protocol_context.comment("You have selected >48 samples. Please Enter in only <=48 samples. Reload the protocol script and correct the sample number!")
#continues on with actual pipetting code...
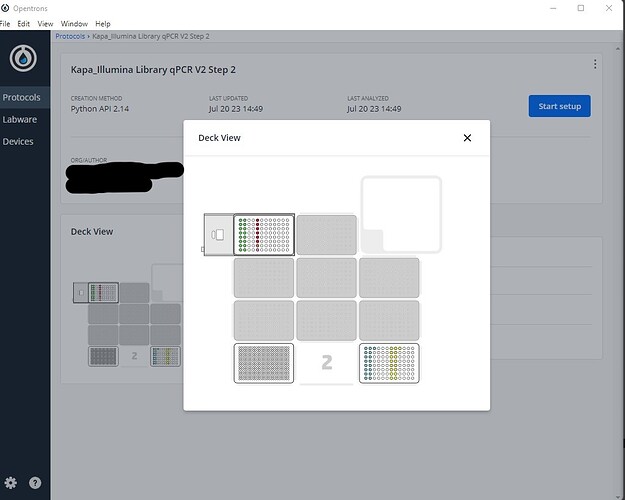
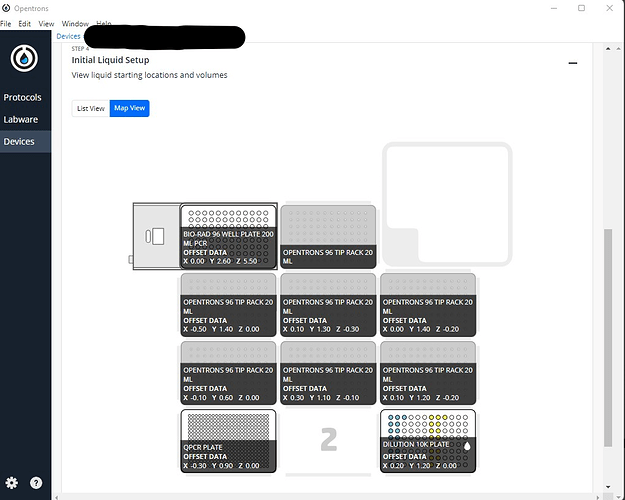
***Note that there does seem to be a bug with labeling of liquids on labware that has been loaded onto the Temperature Module. In the above method, I have created 6 Liquids in this method; 2 for a sample dilution plate, and 4 that go on a Reagent Plate. The method simulates fine, and in the Deck View , I can see all wells labeled correctly.
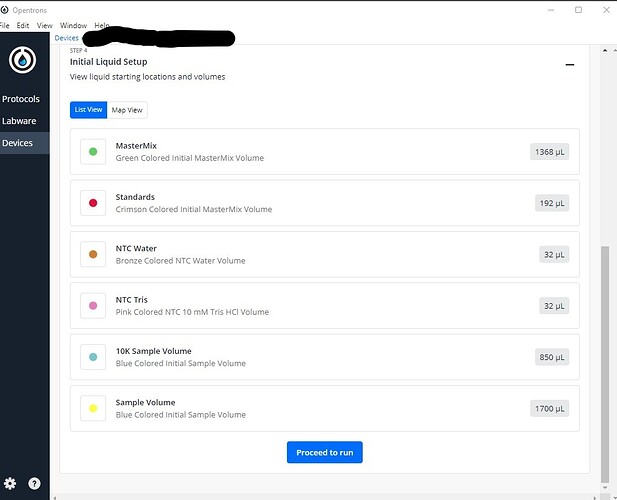
However, once I “Start Setup”, and the protocol is fully analyzed, the Liquids are no longer labeled in the “Map View” of the Step:4 Initial Liquid Setup Section.
The Liquids show up properly in the “List View” of the Step:4 Initial Liquid Setup Section.
When I reported this bug to Opentrons, they were able to replicate the issues I had. I haven’t tried to replicate this on labware that sits on other modules either. The estimation was that a fix would not be available for ~3 months or so.
I hope this helps someone out in the future!